|
|
|
We are thrilled to declare that the unwavering dedication of our research team, led by Yoomin Jeon under Prof.Howard Lee's supervision has culminated in the successful approval of the first patent by CCADD!
This patent covers our novel machine learning-based dimensionality reduction model designed to revolutionize the assessment of clinical trial feasibility. By leveraging electronic medical records to identify common sensitive eligibility features, the research team aimed to significantly improve the efficiency and precision of clinical trial feasibility assessment. This accomplishment not only marks a significant milestone of our research project, but also paves the way for enhanced clinical trial practices in the future. |
|
|
|
|
The 2019 fall conference of The Korean Society of Medical Informatics (KOSMI), organized by Kyungpook National University Hospital (KNUH), was held November 8-9, 2019, at the Global Plaza, Kyunpook National University, Daegu. Prof. Howard Lee, Yoomin Jeon and Junsik Moon participated in the conference.
The conference presented 19 symposia, 6 oral presentation sessions of pre-selected paper submissions, and 3 tutorials in healthcare information, pharmaceutical and big data research which focused on analysis, application and standardization of data based on artificial intelligence (AI) and machine learning. Sharing research ideas and analysis techniques, the attendees had an in-depth discussion on emerging issues and utilization in the fields of medical informatics.

![]()
![]() On the second day, Prof. Lee chaired one of the oral presentation sessions entitled "New Drug and Pharmacovigilance", where he stressed full understanding of the research domain by communicating with experts in each research field. On the other hand, Yoomin presented two posters titled 'A Dimensionality Reduction Model to Increase the Efficiency and Accuracy of Clinical Trial Feasibility Assessment Using Electronic Medical Records' and 'A CDM-based Clinical Trial to Evaluate the Efficacy and Safety of SGLT2 Inhibitors as Add-on Treatment in Patients with Type 2 Diabetes Mellitus on Metformin, Sulfonylurea and/or DPP-4 Inhibitors'. Yoomin's poster presentations were well received. On the second day, Prof. Lee chaired one of the oral presentation sessions entitled "New Drug and Pharmacovigilance", where he stressed full understanding of the research domain by communicating with experts in each research field. On the other hand, Yoomin presented two posters titled 'A Dimensionality Reduction Model to Increase the Efficiency and Accuracy of Clinical Trial Feasibility Assessment Using Electronic Medical Records' and 'A CDM-based Clinical Trial to Evaluate the Efficacy and Safety of SGLT2 Inhibitors as Add-on Treatment in Patients with Type 2 Diabetes Mellitus on Metformin, Sulfonylurea and/or DPP-4 Inhibitors'. Yoomin's poster presentations were well received.

Through the conference, the three members of CCADD were delighted to get introduced to the leading academicians and industry experts in the field of medical informatics. Furthermore, Prof. Lee and two CCADD members had a fun discussion that helped them look deeply into their current activities and direction of future research. Their collected insights out of the conference will be further discussed at the upcoming year-end workshop in early December. |
|
|
|
|
On April 24, 2019, Prof. Howard Lee and 5 CCADD members presented a seminar at the Laboratory of Genome Epidemiology and Health Big Data (G-Epi), Graduate School of Public Health at Seoul National University. It was a return visit in response to G-Epi's one 10 months ago. G-Epi (PI: Joohon Sung) focuses on the data analysis of genetic, demographic, physiological, and environmental factors of complex diseases. G-Epi uses epidemiological, statistical, and bioinformatics approaches to analyze large-scale omics data.
In the seminar, Prof. Howard Lee introduced G-Epi to one of the CCADD's current Artificial Intelligence (AI) research projects, entitled 'A dimensionality reduction model to increase the efficacy and accuracy of clinical trial feasibility assessment using electronic medical records.' Hyun A Lee and Dr. Jeong-An Kim also spoke about their research projects, a PBPK modeling analysis of an acid-reducing agent under clinical development and a pharmacoepigenomic analysis of Tacrolimus, respectively. The seminar was a great opportunity for both labs to share their research experiences, ideas, and future perspectives. G-Epi and CCADD are looking forward to closely working together in various research projects in the future.
|
|
|
|
|
Prof. Howard Lee and Yoomin Jeon attended the 2018 Conference on Empirical Methods in Natural Language Processing (EMNLP 2018) held at the Square Meeting Center in Brussels from October 31st through November 4th, 2018. EMNLP is one of the top-tier conferences in the area of natural language processing organized by SIGDAT, the Association for Computational Linguistics (ACL) special interest group on linguistic data and corpus-based approaches to NLP. With the current artificial intelligence and machine learning boom from a variety of disciplines, this year's conference had a record-breaking number of submissions (2,231) and 2,500+ attendees, a 48% increase compared with EMNLP 2017. Of all the long and short papers submitted, only 549 papers were accepted, which can be found in the EMNLP proceedings. The conference included two days of workshops, followed by three days of main conference sessions including long and short presentations, tutorials, and demos. Researchers and practitioners from around the world gathered to share their latest research findings and emerging trends in machine learning for NLP, machine translation, language models, text mining, information extraction, and many more.

In the 'Writing Code for NLP Research' tutorial, researchers from Allen AI Institute shared their best practices learned from designing the development of the Allen NLP toolkit, a PyTorch-based open-source library for deep learning NLP research. It was an excellent tutorial to learn practical advice on writing research code and to recognize the importance of having good quality codes. In the 'Deep Latent Variable Models of Natural Language' tutorial, deep latent variable models were covered, which are emerging as a useful tool in NLP applications. Mainly, three types of latent variable models (discrete, continuous, and structured) and inference strategies (exact gradient, sampling, and conjugacy) over the latent variables were examined.

There were a number of presentations/papers in the clinical domain. One of the interesting papers, titled 'emrQA: A Large Corpus for Question Answering on Electronic Medical Records', proposed a novel semi-automated framework to generate domain-specific large-scale question answering (QA) corpus by leveraging expert annotations on clinical notes for various NLP tasks from the i2b2 challenge datasets. For medical QA on EMR, they released 400K+ question-answer evidence pairs and 1M question-logical forms, a representation useful for generating corpus. Generating patient-specific QA from an EMR is an important NLP task to process a natural language question and provide a human level accuracy answer. Among many datasets that were presented at the conference, a clinical domain dataset MedNLI was included. MedNLI is a Natural Language Inference (NLI) dataset on the medical history of patients annotated by physicians, which we can later refer in our research.

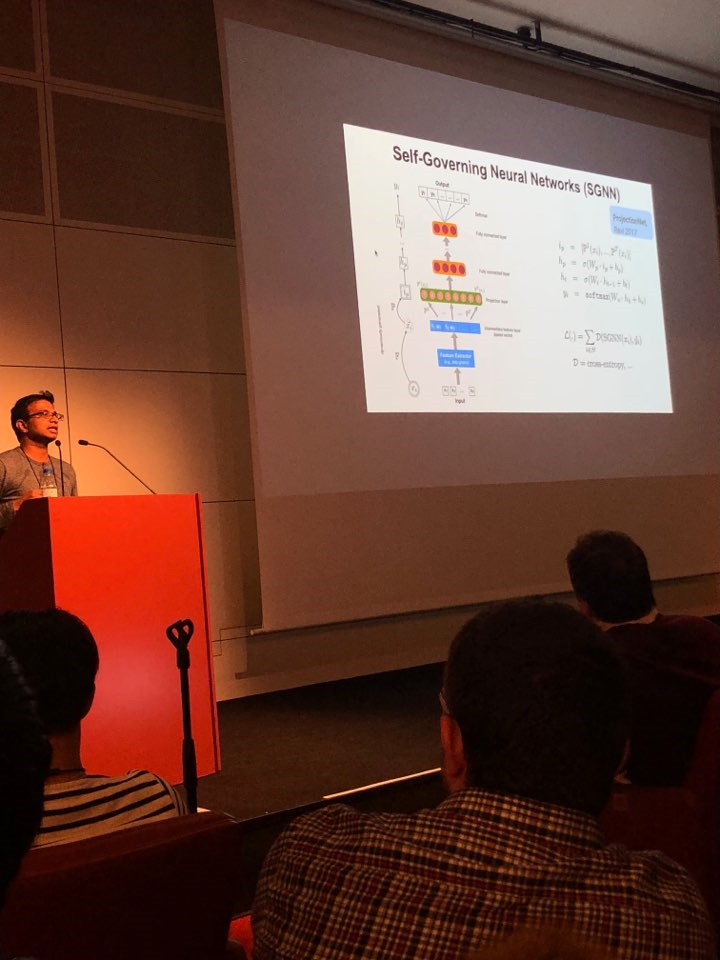
In a paper presented by Google Research, titled ' Self-Governing Neural Networks for On-Device Short Text Classification', researchers described on-device Self-Governing Neural Networks (SGNNs) that achieve state-of-the-art results in dialog related tasks. SGNN uses locality sensitive hashing, a technique for dimensionality reduction in data for clustering. As a result, the embedding layer for their on-device architecture has dropped to 300K parameters compared to other methods having millions of parameters. The proposed method is useful for text classification applications that significantly save storage cost and improve computational performance.
As newcomers to the field focusing on data-driven approaches to NLP, this conference was challenging but worthwhile to see cutting-edge research in NLP. We sure took some inspirations from the conference for our new research to develop a dimensionality reduction model for assessing clinical trials feasibility. We hope to attend the next year's EMNLP conference in Hong Kong!
Okay, below are two cuts of the so-called "Beauty and the Beast." Funny, isn't it?
|
|
|
|
|
Five CCADD members (Prof. Howard Lee, Dr. Soohyun Kim, Yoomin Jeon, Tae Chung, Serin Lee) have attended the Fall conference of The Korean Society of Medical Informatics (KOSMI) held at Chonbuk National University on November 22-23, 2018. The theme of the conference was 'Real World Data to optimize clinical trials', which reflects a growing role and value of big data and artificial intelligence for clinical trials. Over 700 people from academia, public institutions, and the pharmaceutical industry gathered to share their experiences, practices, insights, and perspectives on the more active use of data and information sciences in clinical trials.
Prof. Howard Lee spoke about AI-based patient recruitment in clinical trial in an invited session, entitled 'A Dimensionality Reduction Model to increase the Efficiency and Accuracy of Clinical Trial Feasibility Assessment using Electronic Medical Records(EMR)'. Prof. Lee emphasized the application of AI technology to clinical trial feasibility such as feature selection algorithms in machine learning to identify common sensitive eligibility features (cSEFs). Yoomin Jeon and Tae Chung presented a poster at the conference too. Yoomin's poster was about the actual application of what Prof. Lee introduced in his talk. Tae examined the extent of mapping of ICD-10 to MeSH to crosswalk and synchronize different terms collected from different data sources. (Analysis of agreement status between the diagnostic code of MeSH and ICD-10).
Meanwhile, Dr. Soohyun Kim led a tutorial session, where she taught several essential statistical methods for biomedical informatics research using R.
For more details, you may want to contact Yoomin Jeon or Tae Chung. |
|
|
|
|
Prof. Howard Lee has been recently awarded a government grant from the Small Grant Exploratory Research (SGER) Program through the National Research Foundation of Korea or NRF to develop a model for assessing clinical trials feasibility. The total research funding was $223,000 over a 3 year period (11/01/18 to 10/31/21), which is equivalent to 250 million Korean Won. After a year of an initial stage of research, funding may be continued for additional two more years based on satisfactory progress. The title of Prof. Lee's proposed research plan was "A Dimensionality Reduction Model to increase the efficiency and accuracy of clinical trial feasibility assessment using electronic medical records". This research aimed to develop a machine learning-based dimensionality reduction model to identify the common sensitive eligibility features (cSEFs) that could increase the efficiency of clinical trials feasibility in the presence of intended patient population characteristics. Feature Selection algorithms will be used to enhance the performance of this machine learning model.
|
|
|
|
|
Four CCADD members attended the SNU-Hokkaido University joint symposium held in Sapporo, Japan. On the second day of the symposium, professors and students of the Graduate School of Convergence Science and Technology of Seoul National University met members of the Graduate School of Information Science and Technology of Hokkaido University and shared their research ideas, experiences, and results. The participants from the two universities introduced their research progress on a variety of topics, varying from computer-human interaction study to the development of new drug entities using machine learning technology.
Dr. Soohyun Kim of CCADD gave an oral presentation, where she discussed if therapeutic drug monitoring (TDM) of vancomycin is cost-effective in elderly patients. According to Dr. Kim, the cost-effectiveness of vancomycin in this population might not be so cost-effective as was previously anticipated, but could be cost-effective in patients admitted to the intensive care unit. In the following poster session, Dr. Jeong-An Gim presented the results of a pharmacoepigenomic research (Difference in DNA methylation and pharmacokinetics parameters of tacrolimus between two periods of a bioequivalence test with tacrolimus). Dr. Kim tried to provide a basis for tacrolimus pharmacoepigenomics and may provide a clue to the different dose of tacrolimus through his research. Yoomin Jeon, a PhD student at CCADD, introduced a new research project, entitled "A Dimensionality Reduction Model to Increase the Efficiency and Accuracy of Clinical Trial Feasibility Assessment Using Electronic Medical Records." Yoomin’s poster was associated with a new grant CCADD received recently from the National Research Foundation (PI: Prof. Howard Lee). Another PhD student, Siun Kim, presented his research with Dr. Soohuyn Kim about the validity of the equivalence margin in the phase 3 trial with CT-P13, an Infliximab biosimilar. He had an opportunity to discuss the implication and the future plan of the study with researchers of Hokkaido University.
Those attending the symposium from CCADD were able to meet with many young Japanese professionals and researchers in various fields. They look forward to seeing them again at a joint symposium at Seoul National University next year.




|
|
|
|
|
Artificial Intelligence (AI) is a discipline of computer science that studies the ways to mimic and reproduce human intelligence processes such as learning, knowledge representation, decision-making and reasoning by machines. Several AI-based approaches have been applied to drug discovery and development to increase the efficiency while reducing the time and cost, resulting in a mix of success and failure.
Clinical trials are an important research tool to determine the safety and efficacy of the drug in humans, which s an indispensable knowledge for physicians and patients for the best and optimal care. With the advent of digital health care technology, the fragmented nature of clinical data capture in the previous era is about to change and the way that clinical trials are performed can be also revolutionized. In particular, digital healthcare technology implemented in wearable devices and smart phones has enabled uninterrupted continuous data collection from patients in a real-life setting.

Furthermore, there has been a growing demand for customized, user-friendly digital healthcare platforms that serve each patient’s specialized needs to identify and locate a clinical trial [s]he may be eligible for. Patient recruitment is one of the most expensive, time-consuming, and inefficient steps in any clinical trials. What makes the matter worse is that eligibility assessment is manually conducted by humans (i.e., physicians or study coordinators) d on eyeballing of large amount of patient records and related information. To overcome this drawback, AI-based deep learning algorithms can help match eligible patients to a specific clinical trial and recruiting them into it. Therefore, AI can turn lengthy, laborious, and complex procedures of patient eligibility assessment into several quick and easy clicks on a machine-learning system.
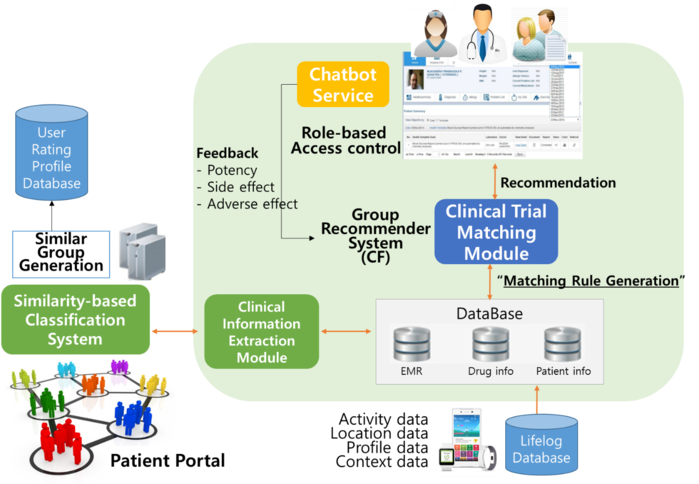
Likewise, it is almost impossible for an average patient, who is lacking the domain knowledge, to find a list of potential clinical trials that [s]he might be eligible for without his/her physician’s help. A simpler, but more efficient, way may be to leverage the utility of AI to identify an appropriate list of clinical trials d on patient’s diagnosis, disease stage, severity and conditions, geographical location, and even personal preferences. Chatbot and the voice-activated assistant can play an important role too in this process.
CCADD has focused on the application of AI technology to clinical drug development, particularly the operational aspects of clinical trials such as eligibility assessment and patient recruit. With the advance of information and communication technology, AI-approach can increase the efficiency of patient recruitment process by providing automated eligibility screening. To analyze heterogeneous patient data, a variety of machine learning algorithms can be used to assess whether a certain patient meets the eligibility criteria, on his/her age, gender, stage of disease, medical history, and clinical conditions. Additionally, dynamic deep neural network can be also used to select clinical features from the electronic medical records (EMR) for eligibility screening. Topic modeling is another helpful tool. To enhance EMR usage for clinical trials, the two most important clinical trials' resources - information on the potential pool and clinical trials being conducted (or have been conducted) - should be fully integrated. By the year 2018, CCADD has successfully developed a clinical trial resource integration system named 'AI-based Clinical Trial Resource Information System' or ACTRiS. What makes ACTRiS unique is its active employment of state-of-the-art user interface (UI) & user experience (UX) technologies, and implementation fo AI technologies. This integrated system will provide a sound basis to design clinical trials that are feasible and practical to perform.
CCADD is currently working on a research project named 'A Dimensionality Reduction Model to increase the efficiency and accuracy of clinical trial feasibility assessment using electronic medical records'. This research aimed to develop a machine learning-basedimensionality reduction model to select the discriminant subset of eligibility features, which adequately returns a sufficient number of eligible patients. This algorithm has the potential to significantly increase the efficiency and performance standard of the traditional approach for patient eligibility screening, contributing to better and more economic conduct of clinical trials.
Also, since spring 2019, CCADD has participated in the three-year project named ' Development an AI-model to predict and evaluate drug-basedrug interactions (DDIs)'. In this project, CCADD is responsible for collecting and curating drug-food interaction information (DFI) from publicly available research papers. Now, we are developing NLP models that recognize name entities of drug and food and classify whether a sentence in an abstract of scientific literature contains a valid DFI information or not. Also, we plan to validate a developed system by verifying whether predicted drug-basedrug pairs as having DDI cause some meaningful change of safety and efficacy of victim drug using a Common Data Model (CDM) of SNUH.
|
|
|
|
|
Many of CCADD members have attended the Spring Conference of The Korean Society of Medical Informatics (KOSMI), held at Samsung Medical Comprehensive Cancer Center on June 14-15, 2018. Reflecting the recent drastic increase in the demand and importance of data science in health sciences, the theme of the conference was 'Evolving Data for Better Health'. More than 600 researchers, practitioners, and vendors in health informatics gathered together to share their knowledge, experience, results of research, and expertise through a series of keynote speeches, symposia, oral/poster presentations, and exhibitions.
Four abstracts by CCADD were accepted for poster presentation. Yoomin Jeon proposed a new approach to transform clinical trials eligibility criteria (EC, free-text) into structured data using the OMOP-CDM(Feasibility of Using the OMOP Common Data Model to Assess Clinical Trial Feasibility). She found that >75% of the EC could be mapped by the proposed approach, and the most important data group was condition.
Serin Lee, a student intern, gave her poster presentation entitled 'Dimensionality Reduction Model for Common Eligibility Criteria to effectively use Electronic Medical Record: A Pilot Study.' She proposed a dimensionality reduction model for common eligibility features (CEFs) based on ontology and semantic features of EC to reasonably compose the concept sets.
Dr. Soohyun Kim proposed a new type of command data model for clinical trials (Clinical Trial Common Data Model, CT-CDM) to support decision-making processes in clinical trials, entitled 'CDISC-based Common Data Model for Developing an Integrated Decision-Support System in Clinical Trials'. This command data model fully complies with the standards of Clinical Data Interchange Standards Consortium(CDISC) as a global, platform-independent data standard guaranteeing system interoperability.
Lastly, but not the least, Dr. Jeong-An Gim shared his research findings: Application of Machine Learning Technology to Classify Healthy Subjects into Different Pharmacokinetic Exposure Groups to Tacrolimus. Using the decision-tree approach employed in R, Dr. Gim aimed to identify SNPs that are associated with the exposure to tacrolimus. The rs776746 in the CYP3A5 gene was the most influential mutation.
More detailed research findings can be downloaded below.
Much to everyone's delighted surprise, Serin Lee won an external solid-state drive of 1TB at the raffle draw! Since Serin will graduate in August, the prize was certainly a nice present.

|
|