|
|
|
Prof. Howard Lee and Yoomin Jeon attended the 2018 Conference on Empirical Methods in Natural Language Processing (EMNLP 2018) held at the Square Meeting Center in Brussels from October 31st through November 4th, 2018. EMNLP is one of the top-tier conferences in the area of natural language processing organized by SIGDAT, the Association for Computational Linguistics (ACL) special interest group on linguistic data and corpus-based approaches to NLP. With the current artificial intelligence and machine learning boom from a variety of disciplines, this year's conference had a record-breaking number of submissions (2,231) and 2,500+ attendees, a 48% increase compared with EMNLP 2017. Of all the long and short papers submitted, only 549 papers were accepted, which can be found in the EMNLP proceedings. The conference included two days of workshops, followed by three days of main conference sessions including long and short presentations, tutorials, and demos. Researchers and practitioners from around the world gathered to share their latest research findings and emerging trends in machine learning for NLP, machine translation, language models, text mining, information extraction, and many more.

In the 'Writing Code for NLP Research' tutorial, researchers from Allen AI Institute shared their best practices learned from designing the development of the Allen NLP toolkit, a PyTorch-based open-source library for deep learning NLP research. It was an excellent tutorial to learn practical advice on writing research code and to recognize the importance of having good quality codes. In the 'Deep Latent Variable Models of Natural Language' tutorial, deep latent variable models were covered, which are emerging as a useful tool in NLP applications. Mainly, three types of latent variable models (discrete, continuous, and structured) and inference strategies (exact gradient, sampling, and conjugacy) over the latent variables were examined.

There were a number of presentations/papers in the clinical domain. One of the interesting papers, titled 'emrQA: A Large Corpus for Question Answering on Electronic Medical Records', proposed a novel semi-automated framework to generate domain-specific large-scale question answering (QA) corpus by leveraging expert annotations on clinical notes for various NLP tasks from the i2b2 challenge datasets. For medical QA on EMR, they released 400K+ question-answer evidence pairs and 1M question-logical forms, a representation useful for generating corpus. Generating patient-specific QA from an EMR is an important NLP task to process a natural language question and provide a human level accuracy answer. Among many datasets that were presented at the conference, a clinical domain dataset MedNLI was included. MedNLI is a Natural Language Inference (NLI) dataset on the medical history of patients annotated by physicians, which we can later refer in our research.

In a paper presented by Google Research, titled ' Self-Governing Neural Networks for On-Device Short Text Classification', researchers described on-device Self-Governing Neural Networks (SGNNs) that achieve state-of-the-art results in dialog related tasks. SGNN uses locality sensitive hashing, a technique for dimensionality reduction in data for clustering. As a result, the embedding layer for their on-device architecture has dropped to 300K parameters compared to other methods having millions of parameters. The proposed method is useful for text classification applications that significantly save storage cost and improve computational performance.
As newcomers to the field focusing on data-driven approaches to NLP, this conference was challenging but worthwhile to see cutting-edge research in NLP. We sure took some inspirations from the conference for our new research to develop a dimensionality reduction model for assessing clinical trials feasibility. We hope to attend the next year's EMNLP conference in Hong Kong!
Okay, below are two cuts of the so-called "Beauty and the Beast." Funny, isn't it?
|
|
|
|
|
Five CCADD members (Prof. Howard Lee, Dr. Soohyun Kim, Yoomin Jeon, Tae Chung, Serin Lee) have attended the Fall conference of The Korean Society of Medical Informatics (KOSMI) held at Chonbuk National University on November 22-23, 2018. The theme of the conference was 'Real World Data to optimize clinical trials', which reflects a growing role and value of big data and artificial intelligence for clinical trials. Over 700 people from academia, public institutions, and the pharmaceutical industry gathered to share their experiences, practices, insights, and perspectives on the more active use of data and information sciences in clinical trials.
Prof. Howard Lee spoke about AI-based patient recruitment in clinical trial in an invited session, entitled 'A Dimensionality Reduction Model to increase the Efficiency and Accuracy of Clinical Trial Feasibility Assessment using Electronic Medical Records(EMR)'. Prof. Lee emphasized the application of AI technology to clinical trial feasibility such as feature selection algorithms in machine learning to identify common sensitive eligibility features (cSEFs). Yoomin Jeon and Tae Chung presented a poster at the conference too. Yoomin's poster was about the actual application of what Prof. Lee introduced in his talk. Tae examined the extent of mapping of ICD-10 to MeSH to crosswalk and synchronize different terms collected from different data sources. (Analysis of agreement status between the diagnostic code of MeSH and ICD-10).
Meanwhile, Dr. Soohyun Kim led a tutorial session, where she taught several essential statistical methods for biomedical informatics research using R.
For more details, you may want to contact Yoomin Jeon or Tae Chung. |
|
|
|
|
Prof. Howard Lee and Hyun A Lee attended the 9th American Conference on Pharmacometrics (ACoP9), held October 7th to 10th, 2018, at the Loews Coronado Bay Resort near San Diego, CA, USA. ACoP is the annual scientific meeting of the International Society of Pharmacometrics (ISoP), an organization comprised individuals from around the globe from various backgrounds in pharmaceutics, engineering, statistics, and mathematics, who are passionate about advancing and promoting the field of pharmacometrics. The theme of ACoP9, "Modeling without Bounds", means "internationalization/collaboration" and "fusion/integration" of pharmacometrics groups, methods, and tools among the fields of pharmacometrics and systems pharmacology, with more than 1,000 members from over 30 countries. Consistent with this theme, inspiring and innovative talks by the Keynote (Mr. Walter Woltosz, Simulations Plus) and the State of Art ( Prof. Andrew Lo, Massachusetts Institute of Technology) speakers were presented. The titles of their speeches were "Rocket motors, Stephen Hawking, and drug development" and "p-Values vs. Patient values: An Analytic Perspective", respectively. In addition, the pre-conference program on "Digital Health Technology, Machine Learning and Artificial Intelligence: The Future of Clinical Trials and Informative Therapeutic Decisions?" highlighted the latest innovations and clinical applications of digital health technologies. The conference program included >100 invited lectures/presentations and over 300 posters, tutorials, pre- and post-meeting workshops, and abundant networking opportunities with attendees from around the world.
Hyun A Lee presented a poster at the meeting that detailed the development of a physiologically based pharmacokinetic (PBPK) model of YH4808, a novel potassium-competitive acid blocker to treat gastric acid related disease. In this study, the mechanistic cause of the decreased exposure YH4808 after multiple oral administration particularly at higher doses could be identified using the PBPK model.
In addition, you may want to contact Hyun A for her poster. |
|
|
|
|
Two scientists from Samsung SDS, Mr. Jaeho Yang, Principal Engineer (Sr. Expert), IoT Lab, and Dr. Sunghoon Joo, PhD, Senior Engineer, Advance Research Lab, visited CCADD on August 24, 2018 to have a joint seminar. In this joint seminar, Prof. Howard Lee gave an overview of the research projects CCADD has performed in the area of applying the artificial intelligence (AI) technology to clinical drug development, entitled with 'AI-enriched Clinical Trials'. Prof. Lee emphasized how AI could streamline various research activities pertaining to clinical trials, one of which is patient enrollment. For example, it is important to know who did what and how before planning any clinical trial, which is the main theme of the grant that the Korea Clinical Trials Global Initiative or KCGI has recently awarded CCADD, named 'AI-based Clinical Trial Resource Integration System or ACTRiS',
Following Prof. Lee's presentation, Mr. Yang introduced CCADD to the core concepts of several AI technologies and their potentials in drug discovery. Recurrent Neural Network (RNN) and autoencoding (AE) were a few examples Mr. Yang put emphasis on during his presentation. After that, Dr. Joo expanded further on several research projects Samsung SDS has conducted to design new molecules for drug discover in silico using advanced AI technologies such as molecular graphs and AE. Collectively, Mr. Yang and Dr. Joo showed how AI could fasten the discovery of new drug candidates that are likely to maintain excellent bio-physico-chemical properties, which may save time, resources, and costs in the whole drug development program.
CCADD and Samsung SDS agreed to continue looking for a cadre of collaboration and joint research opportunities that may benefit both organizations. |
|
|
|
|
Two senior undergraduate students (Left: Hee Chang Lim, Right: Eun Gyung Kim) will be spending the next eight weeks (July-August 2018) at CCADD, participating in the summer internship program at the Graduate School of Convergence Science and Technology, Seoul National University.
Hee Chang is a 4th year undergraduate student at the College of Liberal Studies, Seoul National University. He is majoring in Economics and Electrical Engineering. Hee Chang's research interests include how big data and the use of Artificial Intelligence (AI) can design safer clinical trials. He is looking forward to working as a team member of the CCADD's current research project, named AI-based on Clinical Trial Resource Integration System (ACTRiS).
Eun Gyung is also a senior, currently enrolled in the School of Natural Science, Ulsan National Institute of Science and Technology (UNIST), majoring in Chemistry. She wants to study further on theoretical and computational methods to describe and predict bio-molecular dynamics between proteins, drugs, and other bio-systems. In addition, she has developed a broad interest in computer-aided drug modeling and design.
Let us join welcoming the two amazing student interns, and wishing them a hot summer to achieve many and have fun! |
|
|
|
|
Over the last decade, we have witnessed how technology innovations, such as smartphones and artificial intelligence, can quickly change our lives. The biopharmaceutical industry is not an exception. Since the human genome project was successfully completed, the cost of genetic analysis has rapidly decreased, making genetic analysis more affordable for routine patient care. Furthermore, personalized medicine combined with artificial intelligence technology shed bright light on the possibility of reanalyzing the relationship between patients and diseases.
More extensive development and use of biological agents or biologics d on advanced medical technologies is one of those new trends. To support this notion, it is estimated that almost 50% of research expense by big pharmaceutical companies has been spent in the development of biologics. Biologics currently occupy approximately a quarter of the total pharmaceutical market in terms sales, which is on the sharp rise. For example, seven out of the top 10 best-selling drugs in 2019 were biologics.
However, lay people know little about biologics. This is rather odd because Humulin, the first biopharmaceutical developed by Genentech, was approved by FDA more than 35 years ago (1982). Even OKT3, the first monoclonal antibody drug, was first approved in 1986. Their lack of appreciation on biologics may have something do with the fact that the development and production of biologics involves a variety of modern biological disciplines including molecular biology, to which lay people are not easily accessible.
Based on this understanding, we decided to write an introductory book on biologics that may help lay people understand the core principles of biologics and their follow-on drugs or biosimilars in association with their development, manufacturing, and regulatory implications. We hope that the readers will be able to understand the dynamic changes that are happening in the course of developing biologics and the marketplace. The book was finally published in November 2019.
|
|
|
|
|
Artificial Intelligence (AI) is a discipline of computer science that studies the ways to mimic and reproduce human intelligence processes such as learning, knowledge representation, decision-making and reasoning by machines. Several AI-based approaches have been applied to drug discovery and development to increase the efficiency while reducing the time and cost, resulting in a mix of success and failure.
Clinical trials are an important research tool to determine the safety and efficacy of the drug in humans, which s an indispensable knowledge for physicians and patients for the best and optimal care. With the advent of digital health care technology, the fragmented nature of clinical data capture in the previous era is about to change and the way that clinical trials are performed can be also revolutionized. In particular, digital healthcare technology implemented in wearable devices and smart phones has enabled uninterrupted continuous data collection from patients in a real-life setting.

Furthermore, there has been a growing demand for customized, user-friendly digital healthcare platforms that serve each patient’s specialized needs to identify and locate a clinical trial [s]he may be eligible for. Patient recruitment is one of the most expensive, time-consuming, and inefficient steps in any clinical trials. What makes the matter worse is that eligibility assessment is manually conducted by humans (i.e., physicians or study coordinators) d on eyeballing of large amount of patient records and related information. To overcome this drawback, AI-based deep learning algorithms can help match eligible patients to a specific clinical trial and recruiting them into it. Therefore, AI can turn lengthy, laborious, and complex procedures of patient eligibility assessment into several quick and easy clicks on a machine-learning system.
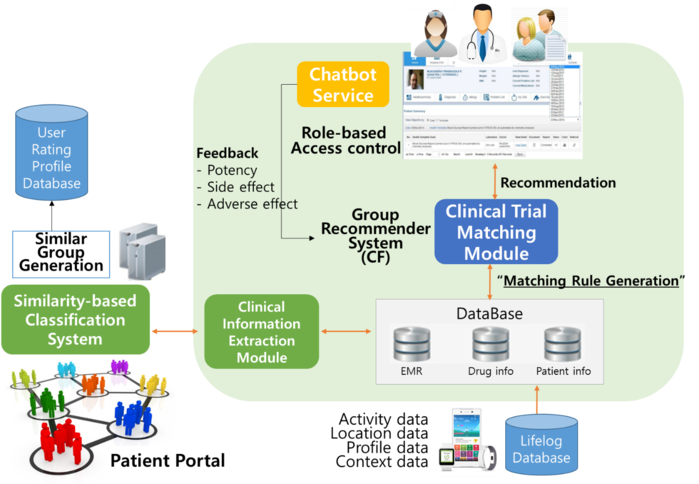
Likewise, it is almost impossible for an average patient, who is lacking the domain knowledge, to find a list of potential clinical trials that [s]he might be eligible for without his/her physician’s help. A simpler, but more efficient, way may be to leverage the utility of AI to identify an appropriate list of clinical trials d on patient’s diagnosis, disease stage, severity and conditions, geographical location, and even personal preferences. Chatbot and the voice-activated assistant can play an important role too in this process.
CCADD has focused on the application of AI technology to clinical drug development, particularly the operational aspects of clinical trials such as eligibility assessment and patient recruit. With the advance of information and communication technology, AI-approach can increase the efficiency of patient recruitment process by providing automated eligibility screening. To analyze heterogeneous patient data, a variety of machine learning algorithms can be used to assess whether a certain patient meets the eligibility criteria, on his/her age, gender, stage of disease, medical history, and clinical conditions. Additionally, dynamic deep neural network can be also used to select clinical features from the electronic medical records (EMR) for eligibility screening. Topic modeling is another helpful tool. To enhance EMR usage for clinical trials, the two most important clinical trials' resources - information on the potential pool and clinical trials being conducted (or have been conducted) - should be fully integrated. By the year 2018, CCADD has successfully developed a clinical trial resource integration system named 'AI-based Clinical Trial Resource Information System' or ACTRiS. What makes ACTRiS unique is its active employment of state-of-the-art user interface (UI) & user experience (UX) technologies, and implementation fo AI technologies. This integrated system will provide a sound basis to design clinical trials that are feasible and practical to perform.
CCADD is currently working on a research project named 'A Dimensionality Reduction Model to increase the efficiency and accuracy of clinical trial feasibility assessment using electronic medical records'. This research aimed to develop a machine learning-basedimensionality reduction model to select the discriminant subset of eligibility features, which adequately returns a sufficient number of eligible patients. This algorithm has the potential to significantly increase the efficiency and performance standard of the traditional approach for patient eligibility screening, contributing to better and more economic conduct of clinical trials.
Also, since spring 2019, CCADD has participated in the three-year project named ' Development an AI-model to predict and evaluate drug-basedrug interactions (DDIs)'. In this project, CCADD is responsible for collecting and curating drug-food interaction information (DFI) from publicly available research papers. Now, we are developing NLP models that recognize name entities of drug and food and classify whether a sentence in an abstract of scientific literature contains a valid DFI information or not. Also, we plan to validate a developed system by verifying whether predicted drug-basedrug pairs as having DDI cause some meaningful change of safety and efficacy of victim drug using a Common Data Model (CDM) of SNUH.
|
|
|
|
|
On April 6, 2018, Prof. Howard Lee gave a special lecture on the principles of clinical trials to visitors from Syntekabio Inc.. They were Dr. Tyson Kim, CEO, Dr. HyunJin Yang, and 4 interns (medical and pharmacy students). Syntekabio Inc. is a South Korean bioinformatics venture firm focusing on genome integration based on big data analysis through the innovative use of artificial intelligence technology. The lecture was followed by an extensive and lively discussion among all the participants, including CCADD members. Tyson Kim also shared his experiences and perspective on the pharmaceutical industry in terms of using bioinformatics technology to streamline drug development. It was a unique opportunity to broaden knowledge beyond the academy. Both CCADD and Syntekabio expect to have more opportunities like this one to advance both party's interests in a collaborative way. |
|
|
|
|
Dr. Kyeong-Ryoon Lee is a research scientist at Korea Research Institute of Bioscience and Biotechnology (KRIBB). Dr. Lee also serves as an Visiting Associate Professor at the Graduate School of Convergence Science and Technology, Seoul National University. He received his PhD in Pharmaceutical Science at Seoul National University with a dissertation, entitled "Neuropharmacokinetic analysis and delivery strategy for drugs targeted to the central nervous system". Dr. Lee's research interests include pharmacokinetic/pharmacodynamic modeling and simulation, artificial intelligence for drug discovery and development, analysis of big data, and model-based translational research in preclinical and clinical studies. |
|
|
|
|
Dr. Jeong-An Gim has joined CCADD on December 1 as a Postdoctoral Researcher at the Graduate School of Convergence Science and Technology, Seoul National University. Prior to joining CCADD, Dr. Gim was a Postdoctoral Researcher at the Life Sciences Department, Ulsan National Institute of Science and Technology (2017), where he played an important role in the Ulsan 10K genome project. Dr. Gim received his master's and doctoral degrees from Pusan National University.
Having a strong background in biological science, Dr. Gim's current research interests include combining advanced computer science technologies such as machine learning and artificial intelligence with genomics and bioinformatics. Dr. Howard Lee, Director to CCADD, welcomed Dr. Gim, saying "With his vast research experience and numerous skill sets in genomics, Dr. Gim will certainly add another layer of opportunities to the existing research capabilities of CCADD." More details about Dr. Gim's past experience and research interests can be found here. |
|
|
|
|
Dr. Howard Lee emphasized data-driven feasibility assessment from the very planning stage of a clinical trial, preferably strengthened and supported by artificial intelligence (AI) technology using the hospital's electronic medical record (EMR) at a KoNECT Forum (November 16, 2017). "Many physicians do not know where a clinical trial is being performed, for which their patients might qualify, and this discouraged the clinicians from talking to their patients about the clinical trials that could benefit them from participating", Dr. Lee explained. Dr. Lee added, "given the drastically increased complexity in the eligibility criteria for patients, EMR-based assessment, particularly coupled with AI, could solve many issues clinical trial investigators are suffering from." CCADD is investigating how AI can be used in clinical trials, particularly for streamlining patient selection and enrollment. |
|
|
|
|
Dr. Yuchae Jung introduced the concept of P-MATCH or Precision Machine Learning AssisTed Clinical Trial Eligibility Assessment using Hospital Records at the 5th Joint Conference on Medical Device between Seoul National University Colleges of Medicine and Engineering and Seoul National University Hospital on November 3, 2017. The title of her talk was 'Application of Artificial Intelligence Technology for Efficient Clinical Trials'. Dr. Jung underlined why artificial intelligence, mainly deep machine learning, can and should be adopted to bring more efficiency to clinical trials, particularly for precise patient enrollment, which allows for patients who are likely to respond to a treatment under testing to have a bigger chance of enrollment. This will also make clinical trials more productive and error-free, Dr. Jung added, because all of the eligibility assessment procedures are algorithm-based using the electronic medical record (EMR).
Dr. Jung's talk was well received, and there were a couple of questions as to what could make P-MATCH different from other approaches such as the IBM Watson for Oncology. Dr. Jung replied that the biggest difference is P-MATCH's natural language processing capability of more than one language (i.e., Korean and English), which often are used in EMR in a haphazard and inconsistent manner.
For anyone who wants to learn more about Dr. Jung's presentation, please contact her. |
|